August 2021 update: Good news! after many hiccups, the Sanger have now completed the sequencing for our samples of the yellow-legged Asian hornet. We’re analysing it right now! Watch this space…
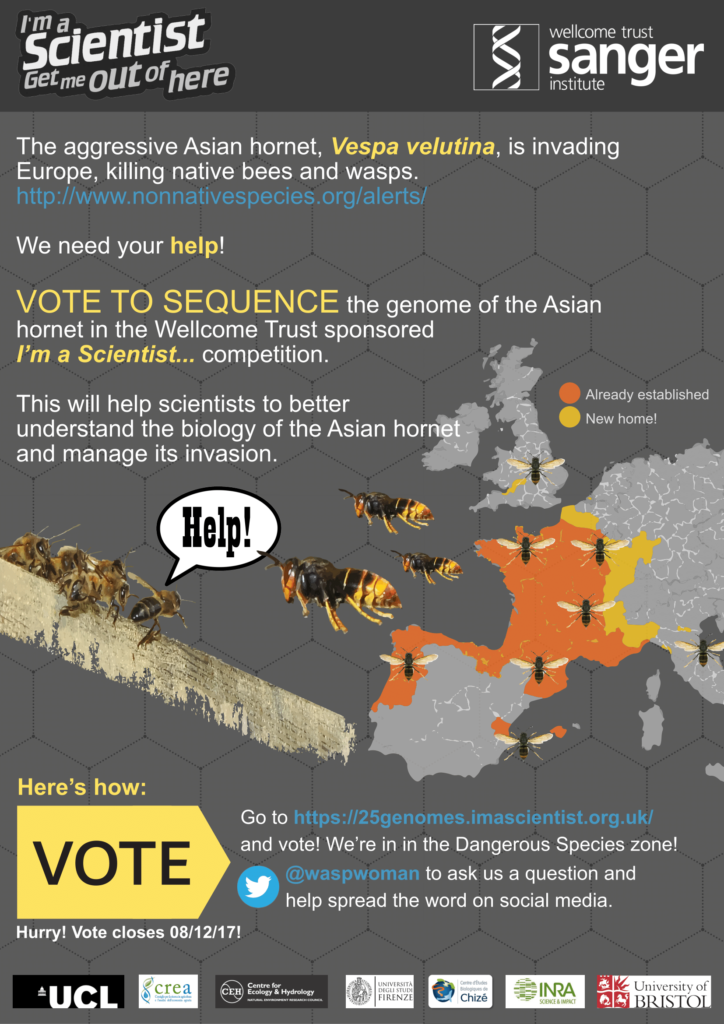
Back in 2017, we pitched for the Asian hornet, Vespa velutina, genome to be sequenced as part of the I’m a Scientist, Get Me Out of Here 25 genomes competition.
The 25 genomes competition celebrated the 25th anniversary of the Wellcome Trust Sanger Institute. Twenty species had already been selected; members of the public, including school children, voted for the final five species in the competition.
We championed the Asian hornet – a dangerous invasive species that poses a huge threat to bee populations in the UK and elsewhere in Europe. Our collaborators include members of: Centre for Genomic Regulation (Spain), University of Natural Resources and Life Sciences (Austria), University of Bristol (UK), Centre for Ecology & Hydrology (UK), University of Florence (Italy), University of Pisa (Italy), University of La Rochelle (France), University of Bordeaux (France), and The Honey Bee and Silkworm Unit, CREA (Italy).
See http://www.nonnativespecies.org/alerts/ for details.
Thanks to the support of the public, Vespa velutina won!!
Sanger made a little film about our lab and why we wanted to sequence this genome.
https://25genomes.imascientist.org.uk/
Here is a summary of our pitch
How will the genome help?
Firstly, we’ll gain insight into the genetic basis of the adaptations that make the Asian hornet such a successful invader. Similar work is already underway on several other invasive species, including the Asian longhorned beetle (Anoplophora glabripennis), and the fruit fly (Drosophila suzukii). We’ll gain better knowledge about the genetic details underlying sex and caste-determination in the Asian hornet, and the kinds of odours and attractants that this species is sensitive to. This knowledge could be used, for instance, to target males and manage populations by limiting reproduction.
Secondly, we’ll be better placed to monitor populations. The genome will provide us with a rich resource with which to develop markers for tracking the invasion. We’ll also be able to better monitor genetic diversity to identify which sub-populations have been or are likely to be more susceptible to certain types of management programme.
Thirdly, we’ll be better placed to consider options for environmental engineering strategies. For instance, in an extreme scenario we might need to look at using biocontrol options or gene editing approaches. One possibility would be to introduce so-called RNA guided ‘gene-drives’ to spread traits like male sterility through invasive populations. This would represent an enhanced version of the sterile insect technique used to manage fly and mosquito populations. Detailed knowledge of the genome would be essential if work of this nature were to be undertaken.
Our poster